 |
| Fig5302 |
 |
| Fig5303 |
Rho-family G proteins are regulated by the three classes of protein; guanine nucleotide exchange factor (GEF), GTPase activating protein (GAP), and guanine nucleotide dissociation inhibitor (GDI). GEF promotes the exchange of GDP with GTP, which results in the binding of the G proteins to their effector proteins. Typical GEF proteins in this family consist of a Dbl-homology (DH) domain, which exhibits GEF activity, and additional domains that mediate interaction with peptides or lipids. The GTP on activated Rho-family G proteins is hydrolyzed in the presence of GAP to resume the GDP-bound inactive state. GDI not only competes with GEF but also holds Rho-family G proteins in the cytoplasm. Therefore, the dissociation of GDI is a prerequisite for the membrane association and activation of Rho-family G proteins. ERM-family proteins, which are involved in cell-to-cell interaction, release Rho from Rho-GDI, translocating Rho from the cytoplasm to the membrane.
 |
| Fig5302 |
 |
| Fig5303 |
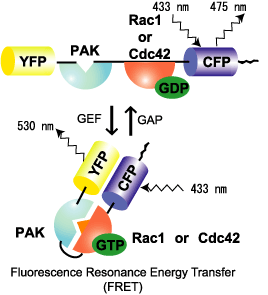
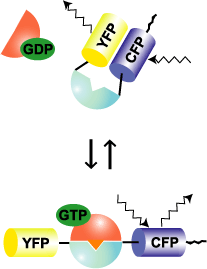
Raichu-Rac and Raichu-Cdc42 use Rac and Cdc42, respectively, as the sensor regions and Pak as the ligand region (Fig. 5302). Upon GTP binding, the effector region of Rac/Cdc42 binds to Pak, which increases FRET from CFP to YFP. In contrast to Raichu-Ras and Raichu-Rap1, Pak was placed before Rac/Cdc42. The reverse order did not work. The probe is anchored to the membrane via the C-terminal region of K-Ras, which is fused to CFP. Since Raichu-Rac1 and Raichu-Cdc42 cannot bind to RhoGDI, these probes cannot monitor the effect of RhoGDI. It should be remembered that the probes monitor the balance between GEFs and GAPs at the membrane. To overcome this insensitivity to RhoGDI, Raichu-CRIB-X was developed (Fig5303). The original idea was proposed by Chalk et al [1]. This type of probe utilizes the nature of GFP to dimerize. By simply sandwiching CRIB with CFP and YFP, we can detect FRET. Fortunately for us, binding of the endogenous Rac or Cdc42 to CRIB was found to inhibit FRET. Thus, this probe can monitor the effect of RhoGDI as well as GEF and GAP. In our preliminary experiment, however, Raichu-CRIB could not detect the change in Rac1 or Cdc42 activity in usual conditions. We attributed this to the excess of probes in the cytosol. Therefore, to reduce the background, we again fused the probe to the C-terminal region of K-ras.
Raichu-Rac1 and Raichu-Cdc42:
These probes are insensitive to RhoGDI. They are designed to monitor the balance between GEFs and GAP activities.
The localization of these probes may not be the same as those of the authentic Rac1 and Cdc42. Recent papers indicate the importance of submicroscopic differences in localization, such as rafts.
Raichu-CRIB-X:
These probes interfere with the physiologic signaling competitively. Indeed, we found that their expression of this probe should be minimal in order to keep the cell in good condition.
Neither of these probes could discriminate between ac and Cdc42. It appears that Cdc42 binds with higher affinity; therefore, the data may be biased for the activity of Cdc42.
HT1080 cells infected with recombinant adenoviruses coding Raichu probes were replated onto a collagen-coated glass-base dish. Beginning 1 hr after replating, CFP, YFP, and differential interference contrast (DIC) images were obtained every 2 min with a time-lapse microscope. A ratio image of YPF/CFP was created to represent FRET efficiency, which correlated with the activities of the G proteins. Video images for 1-hour periods are shown below. As controls, activities of Ras and Rap1 are also shown.
MOV files run on Quick Time Player. MPEG files run on Windowes Media Player
Raichu-Rac1 (MOV, 4 M byte) (MPEG, 0.8 M byte)
Raichu-Cdc42 (MOV, 4 M byte) (MPEG, 0.8 M byte)
Raichu-Ras (MOV, 4 M byte) (MPEG, 0.8 M byte)
The video images show that Rac1 is activated diffusely, with an increasing gradient toward the leading edge. This observation agrees with a previous report by Hahn et al. [2]. In contrast, the Cdc42 activity is more confined to the tip of leading edge.
1. D. L. Graham, P. N. Lowe, and P. A. Chalk. A method to measure the interaction of Rac/Cdc42 with their binding partners using fluorescence resonance energy transfer between mutants of green fluorescent protein. Anal.Biochem. 296:208-217, 2001.
2. V. S. Kraynov, C. Chamberlain, G. M. Bokoch, M. A. Schwartz, S. Slabaugh, and K. M. Hahn. Localized rac activation dynamics visualized in living cells. Science 290:333-337, 2000.